SUMMARY
The Unmet Need: Methods for efficient, reversible integration and excision of large DNA
- Genome engineering is a rapidly advancing field with immense potential for synthetic biology and gene therapy. The ability to precisely modify DNA sequences within living cells is crucial for understanding gene function, correcting genetic defects, and developing novel therapeutic strategies. There is a significant need for technologies that enable the efficient, site-specific, and reversible integration and excision of large DNA sequences into mammalian genomes. Such capabilities are essential for dynamic control over gene expression and for applications requiring temporary genetic modifications.
-
Current genome editing approaches face several limitations. While some methods allow for DNA integration, they often lack the precision or efficiency required for large DNA payloads. A major challenge with existing site-specific recombinases, such as large serine recombinases (LSRs), is their inherent unidirectional nature. These enzymes primarily catalyze integration at specific "attB" and "attP" sites, forming "attL" and "attR" sites, but they are unable to efficiently mediate the reverse excision reaction from these product sites. This unidirectionality restricts the available genomic target sites and prevents the clean, reversible removal of integrated DNA, hindering dynamic genome engineering.
The Proposed Solution: Gene editing system utilizes large serine recombinase-recombination directionality factor (LSR-RDF) fusion proteins for programmable, precise, and reversible large DNA insertions/excision
- The faculty inventor developed novel fusion proteins engineered by LSRs with their cognate recombination directionality factors (RDFs). While LSRs alone catalyze unidirectional DNA integration, these LSR-RDF fusions specifically and efficiently mediate recombination at attL and attR sites. This effectively doubles the available genomic target sites, enabling reversible genome engineering for both integration and clean excision of large DNA sequences. The system is compatible with evolved LSR variants and can be integrated with prime editing for programmable, precise, and reversible large DNA insertions in mammalian cells, with applications in synthetic biology and gene therapy.
- LSRs typically only integrate DNA at attB/attP sites. By fusing LSRs with RDFs, the system gains the unique ability to specifically target attL and attR sites, enabling the reverse excision reaction. Mechanistically, RDF binding redirects a coiled-coil motif within the LSR, altering the architecture of the DNA-bound synaptic complex to control reaction directionality. This unique mechanism allows for a complete and reversible cycle of large DNA integration and removal.
Figure
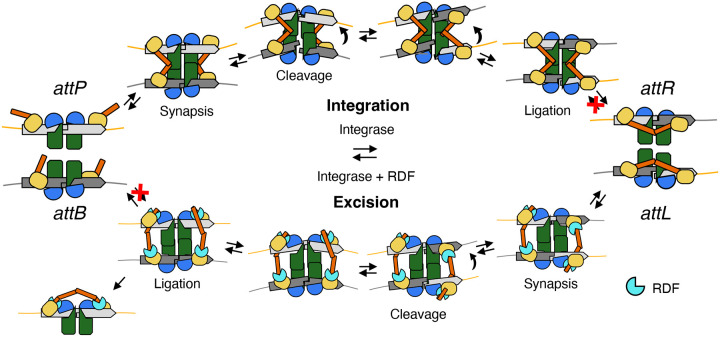
Schematic model of how CC-CC interactions and the RDF mediate directionality control.
Pathways for integrative (upper) and excisive (lower) recombination are diagrammed with the protein domains and DNAs colored as in previous figures. CC subdomains (orange) buttress allowed synaptic complexes by mediating interactions between the upper and lower DNAs, but they can also re-arrange after recombination to form intra-dimer interactions that trap the products and prevent the reverse reaction from occurring (red X’s). The integration pathway is conceptually similar to that proposed by Rutherford and Van Duyne35 but the CC motifs in particular are redrawn to match the experimental data presented here. The RDF redirects the trajectories of the CC subdomains so that a different type of synaptic complex (between attL and attR) and a different product dimer (attP-bound) are stabilized.
ADVANTAGES
ADVANTAGES
-
Enables reversible gene editing, allowing both integration and clean excision of large DNA sequences
-
Doubles the available genomic target sites for large serine recombinases (LSRs)
-
Offers high specificity and efficiency for genome editing with reduced off-target effects
-
Compatible with evolved LSR variants, enhancing flexibility and applicability
-
Integrates with prime editing for precise and programmable large DNA insertions
APPLICATIONS
-
Reversible gene editing
-
Large DNA payload delivery
-
Dynamic synthetic biology
-
Biopharmaceutical production optimization
PUBLICATIONS
August 13, 2025
Proof of concept
Patent Pending
Licensing,Co-development
Weixin Tang