SUMMARY
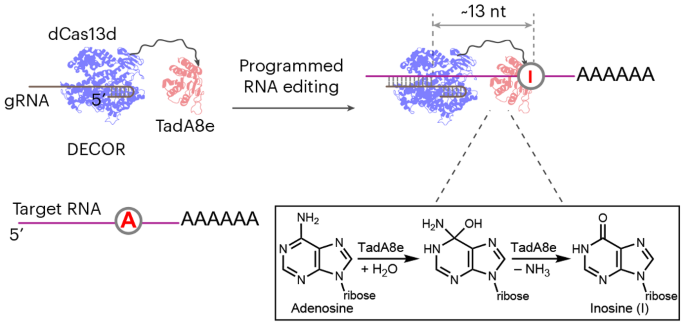
A novel platform, bacterial deaminase-enabled recoding of RNA (DECOR), utilizing an engineered bacterial adenosine deaminase to achieve programmable adenosine-to-inosine (A-to-I) editing on single-stranded RNA.
The Unmet Need: Novel effector proteins for programmed RNA editing
- RNA editing has emerged as a promising therapeutic strategy for addressing genetic diseases. Unlike DNA editing, which permanently alters the genome, RNA editing offers a reversible and dose-dependent approach to modify cellular behavior by targeting RNA transcripts. This flexibility is particularly valuable for diseases requiring precise editing levels or treatments within specific time windows. The potential of RNA editing to treat genetic diseases stems from its ability to correct genetic defects at the RNA level, avoiding permanent modifications to the genome.
-
Current RNA editing approaches primarily rely on the ADAR family of enzymes, which act on double-stranded RNA (dsRNA). However, these ADAR-based methods face limitations. Firstly, the reliance on endogenous ADARs poses a challenge due to their low expression in most adult tissues and their predominant nuclear localization, hindering efficient editing in the cytoplasm. Secondly, the broad target range of ADARs raises concerns about off-target editing, potentially impacting millions of sites in the human transcriptome. Lastly, the inherent sequence preferences of ADARs restrict their editing capabilities to specific adenosine (A) sites, excluding a subset of potential therapeutic targets.
The proposed solution: A CRISPR-based RNA editing platform utilizing a bacterial enzyme to enable precise modifications of RNA sequences in cells
- The faculty inventor developed bacterial deaminase-enabled recoding of RNA (DECOR) which employs an engineered Escherichia coli tRNA adenosine deaminase to perform adenosine-to-inosine (A-to-I) editing at specific sites in the transcriptome. Unlike traditional ADAR-based RNA editing, DECOR targets single-stranded RNA, expanding its editing capabilities beyond existing double-stranded RNA-targeting systems. This system has demonstrated high on-target editing efficiency while significantly reducing off-target effects.
-
DECOR differentiates itself from other RNA editing technologies through its unique features. Firstly, it is the first ADAR-independent A-to-I editing platform, expanding the available tools for programmed RNA editing. Secondly, it exhibits broad compatibility with various RNA-targeting CRISPR systems, enabling flexible and efficient editing.
-
Importantly, DECOR demonstrates a superior safety profile with significantly lower transcriptome-wide off-target effects compared to ADAR-overexpressing systems. This enhanced specificity is further improved in high-fidelity DECOR variants, minimizing unintended edits. Lastly, DECOR's therapeutic potential is highlighted by its ability to remove a disease-causing upstream open reading frame (uORF) in the interferon regulatory factor 6 (IRF6) gene, restoring normal protein expression levels.
FIGURE
 ADVANTAGES
ADVANTAGES
ADVANTAGES
-
First ADAR-independent A-to-I editing platform
-
Minimal off-target effects
-
Broadly compatible with RNA-targeting CRISPR systems
-
Offers a potential solution for programmed C-to-U editing in RNA
APPLICATIONS
- Gene therapy
- Drug discovery
PUBLICATIONS
October 18, 2024
Proof of concept
Patent Pending
Licensing,Co-development
Weixin Tang