SUMMARY
A class of functionally enhanced synthetic transcriptional factor antagonists to modulate the MYC/MAX transcriptional network by direct competition for DNA binding.
The Unmet Need: Transcription factors remain largely untapped as pharmacologic targets due to the challenges in targeting protein–protein and protein–DNA interactions
- Transcription factors (TFs) regulate cellular state by binding specific DNA promoter and enhancer sequences, thereby recruiting transcriptional machinery to activate or repress gene expression. DNA binding is carried out by diverse DNA-binding domains (DBDs), which are shared across large families of TFs and conserved across evolutionary landscapes.
-
Many archetypal DBDs contain an α-helix oriented in the DNA major groove, which requires defined secondary, tertiary and/or quaternary folds for proper binding. Despite the complex three-dimensional structure encoded by DBDs, numerous studies have demonstrated that many DBDs are inherently modular, which permits retention of potent and specific DNA-binding activity when domains are separated from the rest of their parent TF or when grafted onto entirely new proteins. These finding suggest that molecules that mimic the structural features of DBDs could serve as minimal TF mimetics capable of interfering with or replacing endogenous TF activity.
-
Current approaches in TF mimetics require synthesis or expression of long polypeptides, which suffer from low synthetic yields and poor pharmacologic properties, such as protease stability and cellular penetration. Theses physiochemical properties considerably limit their utility as chemical probes or therapeutics.
The proposed solution: Modular synthetic transcriptional repressor platform for non-natural TF mimetics to target TF function through direct modulation of DNA binding
- The faculty inventor developed a synthetic reconstruction of the MAX protein that is proteolytically stable, cell permeable and capable of binding specific DNA recognition sites in living cells. These non-natural, ‘cross-dimer’ synthetic DNA-Binding Domains (sDBDs) recapitulate the cooperative association of individual DNA binding helices, as found in hundreds of known transcription factors, however, their hybrid architecture suggests they should not interfere with protein-protein interactions between endogenous bHLH proteins.
-
In order to access higher ordered protein topology the inventor established a modular, convergent synthetic route that enabled introduction of multiple non-natural stabilizing elements, and in the process identified the necessary structural features required for high affinity and specific DNA recognition. These sDBDs are the first example of a stabilized proteomimetic or “stapled” peptide to encode secondary, tertiary and quaternary structure
FIGURE
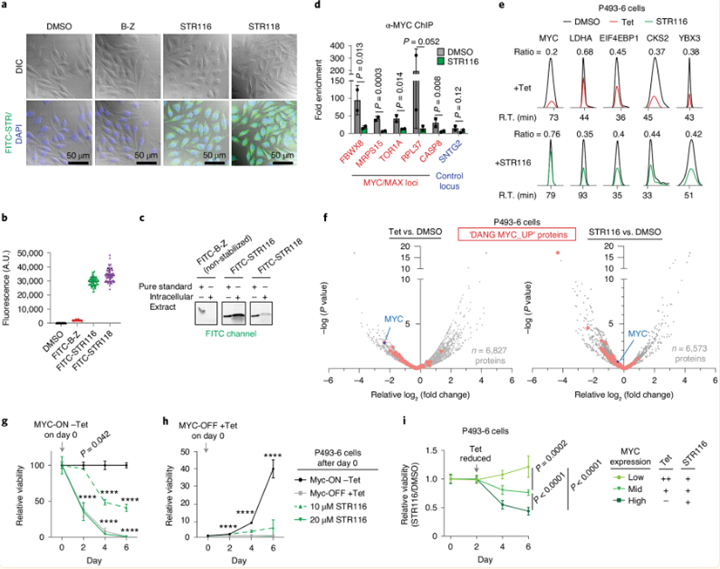
STRs are cell permeable and oppose MYC-dependent gene expression and phenotypes in cells.
a,b, Representative confocal fluorescence microscopy images (a) and quantification of total fluorescence intensity (b) of HeLa cells treated with 5 μM STRs. c, Fluorescence gel analysis of extracted intracellular contents from HeLa cells treated with 1 μM of the indicated FITC-STR. 1 μM fluorescent standard of each FITC-STR is included for quantitative comparison. d, Competitive ChIP–qPCR quantification of endogenous MYC occupancy at control and E-box-containing target genes in P493–6 cells treated with STR116 (20 μM) or vehicle for 24 hours. e, Representative SILAC chromatograms for MYC and known MYC-regulated proteins in P493–6 cells treated with vehicle (DMSO), Tet (0.1 μg ml−1, 72 hours) or STR116 (20 μM, 48 hours). Ratios listed are the treated versus control ratio for each chromatogram. R.T., retention time. f, Volcano plot depictions of the protein expression profiles from Tet-treated and STR116-treated P493–6 SILAC cells. The location of validated MYC targets from the ‘DANG_MYC_TARGETS_UP’ gene set are highlighted in red, with MYC highlighted in blue. P values were calculated using a background-based t-test. g,h, Relative viability of proliferating (cells starting in the MYC-ON state, left) or rescued (cells starting in the MYC-OFF state and then released, right) P493–6 cells treated with vehicle, Tet or STR116 for each timepoint are shown. i, Relative viability ratio of proliferating P493–6 cells comparing STR116 to vehicle-treated cells under low, mid and high MYC expression induced by dose-dependent Tet treatment. Graph in b shows per-cell values (n = 60 cells) with mean and s.d. shown as solid bars. ChIP–qPCR data represent the mean and s.d. of n = 2 biological replicates. Viability plots show mean and s.d. from n = 3 biological replicates. Statistical analyses are by unpaired, two-sided t-test. ****P < 0.0001. A.U., arbitrary units.
ADVANTAGES
ADVANTAGES
- Improved DNA binding affinity
- More selective DNA binding specificity
- Enhanced proteolytic stability
- Ease of chemical synthesis
- Programmable
APPLICATIONS
- Targeted anti-cancer treatment
- Research tool for the study of cancer cell protein expression and regulation
PUBLICATIONS
-
Speltz TE, Qiao Z, Swenson CS, Shangguan X, Coukos JS, Lee CW, Thomas DM, Santana J, Fanning SW, Greene GL, Moellering RE. Targeting MYC with modular synthetic transcriptional repressors derived from bHLH DNA-binding domains. Nat Biotechnol. 2023 Apr;41(4):541-551. doi: 10.1038/s41587-022-01504-x. Epub 2022 Oct 27. PMID: 36302987; PMCID: PMC10392954.
July 25, 2024
Proof of concept
Patent Pending
Licensing,Co-development
Ray Moellering